Following allopolyploid formation, extensive genome evolution occurs, with the eventual loss of many homeologous gene copies. Although this process of diploidization has occurred many times independently, the evolutionary forces determining the probability and rate of gene loss remain poorly understood. Capsella bursa-pastoris is a recently derived allopolyploid, which resulted from a hybridization event between Capsella grandiflora and Capsella orientalis, approximately 100,000-300,000 years ago. There is extensive population differentiation between Asian and European C. bursa-pastoris. However, within Asia, relatively little population structure has been observed to date, and demographic model fitting suggests that colonization occurred from the Middle East within the last 1000 generations associated with a founder event followed by expansion. Despite the lack of evident population structure, Asian populations experience highly divergent environmental conditions. In China, C. bursa-pastoris populations occur in a broad array of complex environmental conditions ranging from subtropical climatic conditions in the south to arid/semi-arid environment in the northwest and boreal climate in the northeast. And C. bursa-pastoris populations distribute from the eastern low-altitude coast to the western Qinghai-Tibetan plateau with altitudes over 4000 m. Given the short timescale within China and relatively limited population structure, this region provides an outstanding opportunity to examine the extent of expression divergence and local adaptation in an allopolyploid over a short evolutionary timescale.
Dr. Hui-Run Huang and her colleagues, from the Research Group of Molecular Ecology (PI: Prof. Xue-Jun Ge) in South China Botanical Garden, and their collaborators Prof. Stephen I. Wright (University of Toronto) and Prof. Martin Lascoux (Uppsala University), have conducted genome and transcriptome sequencing in a broad sample of Chinese accessions of C. bursa-pastoris. The whole genome data revealed three groups of these accessions: an Eastern group from low-altitude regions, a Western group from high-altitude regions and a much more differentiated Northwestern group (Fig. 1). Population differentiation in total expression was limited among closely related populations; in contrast, the relative expression of the two homeologous copies closely mirrors the genome-wide SNP divergence (Fig. 2). Consistent with this, they observed a negative correlation between expression changes in the two homeologues. However, genes showing population genomic evidence for adaptive evolution did not show an enrichment for expression divergence between homeologues, providing no clear evidence for adaptive shifts in relative gene expression. Overall, these patterns suggest that neutral drift may contribute to the population differentiation in the expression of the homeologues, and drive eventual gene loss over longer periods of time.
This research has been published in New Phytologist (https://nph.onlinelibrary.wiley.com/doi/full/10.1111/nph.15299).
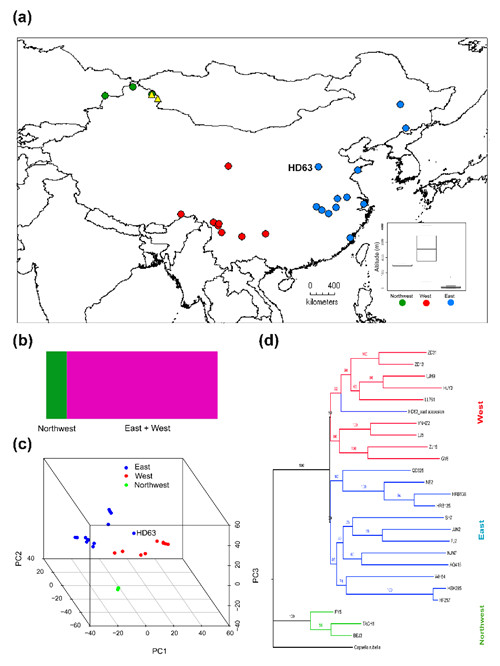
Fig. 1 Geographic distribution and genetic structure of Capsella bursa-pastoris in China. (a) Distribution of the studied populations. (b) Population structure for the C. bursa-pastoris accessions, inferred with fastSTRUCTURE, for K = 2, showing two genetic clusters: the Northwest and the remaining populations. (c) Genetic structure inferred by principal components analyses shows three groups: the Northwest, the West and the East. (d) A maximum likelihood tree inferred by SNPhylo also suggests three groups, with an exception for an eastern accession HD63 which clustered within the West group.
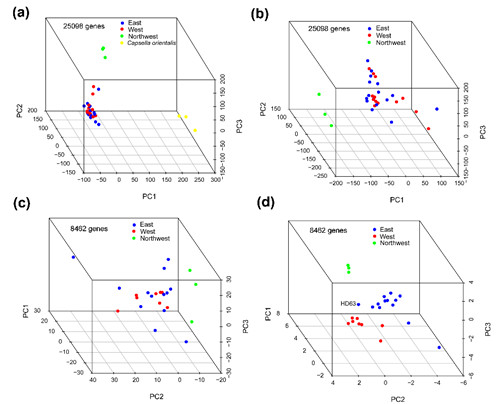
Fig. 2 Principal components analyses (PCA) on expression data sets in Capsella. (a) and (b) PCA on total expression of 25098 genes, where total expression combines estimates across the two homeologues. C. orientalis accessions are included in (a), but excluded in (b). (c) PCA on total expression for a narrow set of genes with sufficient coverage to adequately measure homeologous-specific expression. (d) PCA on homeologue-specific expression.

